The evolution of species by means of natural selection can be modelled in a “tree of life” (ToL) that can be traced backwards in time [1]. At the root of the ToL is a hypothetical being known as the Last Universal Common Ancestor (LUCA) [2,3]. LUCA is in turn the outcome of several evolutionary events distinguished by the primacy of various fundamental components: geochemical prebiotic molecules → nucleotides (nt) + amino acids (aa) → RNA + [aa & peptides] → [RNA & proteins] + aa → [RNA & proteins] + DNA → [DNA + Proteins] + RNA (cf. [4]).
The “RNA world” is the evolutionary stage where autocatalytic and heritable RNA was the prevalent molecule [5,6]; while LUCA content, whose genetic material –DNA– followed the standard genetic code (SGC) appears up to the last phase [7–9].
According to Eigen and Schuster, the primeval genetic code (PGC) was composed of RNA strands following an RNY pattern [puRines (A/G), pYrimidines (C/U), or aNy of these 4 nt] [10,11], whose phenotype are some of the earliest aa produced by prebiotic synthesis [12,13], to wit: glycine (G/Gly), alanine (A/Ala), aspartate (D/Asp), valine (V/Val), serine (S/Ser), threonine (T/Thr), asparagine (N/Asn) and isoleucine (I/Ile); as we can see, the PGC was already redundant with two codons encoding for a single aa.
Two paths that also have a biological correspondence were mathematically derived from the RNY code to the current SGC. On the one hand, modifications to the reading frame brought about by a primitive copy that was less precise than a modern one led to the set RNY+NYR+YRN called “Extended 1” (Ex1); on the other hand, transversions at the first or third base, led to the set RNY+YNY+RNR called “Extended 2” (Ex2). Finally, the extended genetic codes (ExGCs) from both 48-codon collections complemented each other to create the SGC [14,15].
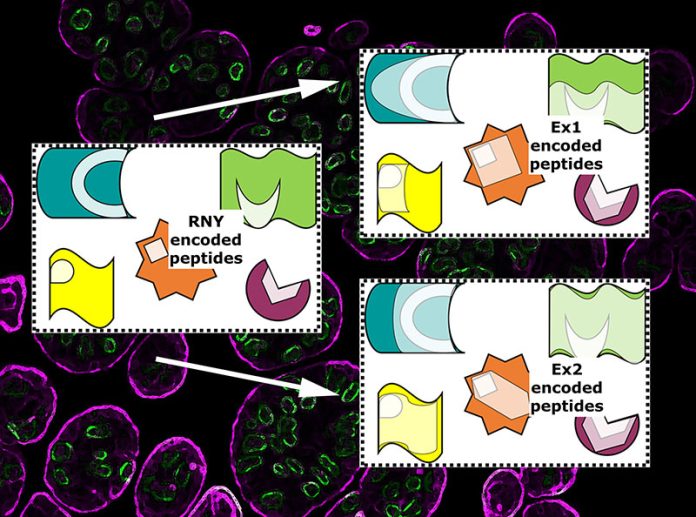
It is feasible to track the evolution of the phenotype using the biologically plausible algebraic derivation of the SGC. We first obtained the RNY, Ex1 and Ex2 genomes of various organisms [14,15], before searching for the corresponding proteome –the complete collection of the proteins of a specific organism– encoded by each of those ancestral genetic codes.
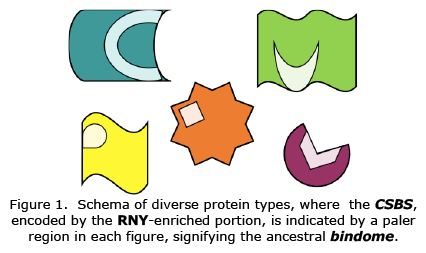
Contrary to expectations, the fundamental protein modules encoded by RNY triplets correspond Cofactor Stabilising Binding Sites (CSBSs) rather than the catalytic sites of modern proteins {Figure 1}, making the primordia of modern proteins, or the Ur-proteome, a bindome for some of the oldest prebiotic molecules formed on the early Earth [16]. Some authors claimed a delineation of the proteins contained in LUCA [17], but our findings show that those motifs were there far earlier. According to our discovery, primordial chemicals were stabilised before they were ever used for catalyses [16].
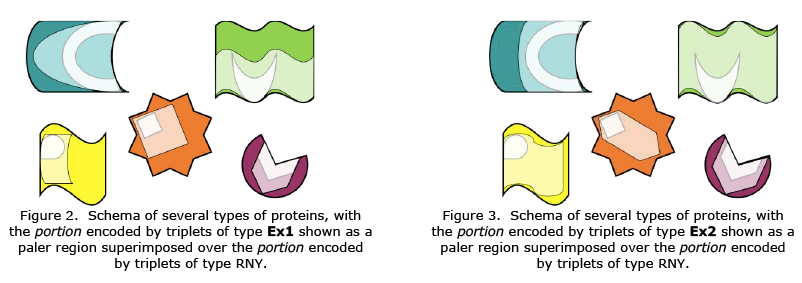
Similarly, the fragments encoded by one or another type of ExGC corresponded to distinct and complementary regions of contemporary proteins {Figures 2 & 3}; those fragments came together and performed similar roles to contemporary ones, constituting an early draught of the full LUCA proteome [18]. Particularly, the Wood-Ljungdahl metabolism key proteins, which were allegedly involved in the metabolism of LUCA but could not previously be located, were in fact
obtained by us [19]. Along with the aforementioned proteins, nearly all of the translation system proteins and several of basal metabolism enzymes, such triose phosphate isomerase, were fully produced prior to LUCA [18]. It is interesting to note that the portions encoded by Ex1 are shorter but more numerous, whereas the ones encoded by Ex2 are few and longer; moreover, just as the ExGCs complement one another to create the SGC, so do the encoded portions [18].
From the early origin of life (OoL) through the emergence of LUCA in the root of the ToL, we believe that the n briefly discussed here enable us to locate, in the form of images of frozen evolutionary moments, remnants of prebiotic and proto-biotic components within present life forms. Finally, the experimental reconstruction of ancient proteins or their ancestors might be guided by this kind of research and shed light on the evolution of biogeochemistry [20–22].
References:
- Darwin, C. On the Origin of Species by Means of Natural Selection, or, The Preservation of Favoured Races in the Struggle for Life /; John Murray,: London :, 1859; Vol. 1859, pp. 1–564.
- Forterre, P.; Gribaldo, S.; Brochier, C. [Luca: the last universal common ancestor]. Med Sci (Paris) 2005, 21, 860–865, doi:10.1051/medsci/20052110860.
- Woese, C. The Universal Ancestor. Proc Natl Acad Sci U S A 1998, 95, 6854–6859.
- Cech, T.R. The RNA Worlds in Context. Cold Spring Harb Perspect Biol 2012, 4, a006742, doi:10.1101/cshperspect.a006742.
- Gilbert, W. Origin of Life: The RNA World. Nature 1986, 319, 618, doi:10.1038/319618a0.
- Gesteland, R.F.; Cech, T.; Atkins, J.F. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World; Cold Spring Harbor Laboratory Press, 2006; ISBN 978-0-87969-739-6.
- Koonin, E.V.; Novozhilov, A.S. Origin and Evolution of the Genetic Code: The Universal Enigma. IUBMB Life 2009, 61, 99–111, doi:10.1002/iub.146.
- Woese, C.R. Universality in the Genetic Code. Science 1964, 144, 1030–1031, doi:10.1126/science.144.3621.1030.
- Woese, C.R. Interpreting the Universal Phylogenetic Tree. PNAS 2000, 97, 8392–8396, doi:10.1073/pnas.97.15.8392.
- Eigen, M. Selforganization of Matter and the Evolution of Biological Macromolecules. Die Naturwissenschaften 1971, 58, 465–523, doi:10.1007/BF00623322.
- Eigen, M.; Schuster, P. The Hypercycle – A Principle of Natural Self-Organization Part C: The Realistic Hypercycle. Naturwissenschaften 1978, 65, 341–369, doi:10.1007/BF00439699.
- Miller, S.L. A Production of Amino Acids Under Possible Primitive Earth Conditions. Science 1953, 117, 528–529, doi:10.1126/science.117.3046.528.
- Bada, J.L. New Insights into Prebiotic Chemistry from Stanley Miller’s Spark Discharge Experiments. Chem Soc Rev 2013, 42, 2186–2196, doi:10.1039/c3cs35433d.
- José, M.V.; Govezensky, T.; García, J.A.; Bobadilla, J.R. On the Evolution of the Standard Genetic Code: Vestiges of Critical Scale Invariance from the RNA World in Current Prokaryote Genomes. PLOS ONE 2009, 4, e4340, doi:10.1371/journal.pone.0004340.
- José, M.V.; Morgado, E.R.; Govezensky, T. Genetic Hotels for the Standard Genetic Code: Evolutionary Analysis Based upon Novel Three-Dimensional Algebraic Models. Bull Math Biol 2011, 73, 1443–1476, doi:10.1007/s11538-010-9571-y.
- Palacios-Pérez, M.; Andrade-Díaz, F.; José, M.V. A Proposal of the Ur-Proteome. Orig Life Evol Biosph 2018, 48, 245–258, doi:10.1007/s11084-017-9553-2.
- Sobolevsky, Y.; Guimarães, R.C.; Trifonov, E.N. Towards Functional Repertoire of the Earliest Proteins. J. Biomol. Struct. Dyn. 2013, 31, 1293–1300, doi:10.1080/07391102.2012.735623.
- Palacios-Pérez, M.; José, M.V. The Evolution of Proteome: From the Primeval to the Very Dawn of LUCA. Biosystems 2019, 181, 1–10, doi:10.1016/j.biosystems.2019.04.007.
- Weiss, M.C.; Sousa, F.L.; Mrnjavac, N.; Neukirchen, S.; Roettger, M.; Nelson-Sathi, S.; Martin, W.F. The Physiology and Habitat of the Last Universal Common Ancestor. Nature Microbiology 2016, 1, 16116, doi:10.1038/nmicrobiol.2016.116.
- Carter, C.W. Urzymology: Experimental Access to a Key Transition in the Appearance of Enzymes. J. Biol. Chem. 2014, 289, 30213–30220,doi:10.1074/jbc.R114.567495.
- Garcia, A.K.; Kaçar, B. How to Resurrect Ancestral Proteins as Proxies for Ancient Biogeochemistry. Free Radical Biology and Medicine 2019,140, 260–269, doi:10.1016/j.freeradbiomed.2019.03.033.
- Hochberg, G.K.A.; Thornton, J.W. Reconstructing Ancient Proteins to Understand the Causes of Structure and Function. Annual Review of Biophysics 2017, 46, 247–269, doi:10.1146/annurev-biophys-070816-033631.