Introduction
The Network of Researchers on the Chemical Evolution of Life (NoRCEL, formally NoR HGT & LUCA) was formed in 2013 with a view to collate and communicate the vastly multidisciplinary aspects of the origin of life. From the epoch of earlier frontiers exploration into life’s origins by scientists such as Charles Darwin (1809–1882) until Stanley Miller (1930–2007) performed his electric discharge experiments, the abiding fact is that the answer to how life began could only be found via an integrative approach. One unique selling point of NoRCEL is that we are actively involved in finding and inviting scientists who are not necessarily directly involved in studies pertaining to the origin of life—e.g., those involved with medical virology, cancer cell research, palaeontology etc. Our members are a mix of practitioners of research into the origin of life and also those who work in areas which may inadvertently produce some germane discovery. A second distinctive feature is that we produce a conference report highlighting our members’ latest research in this particular frontier of science. The fourth NoR CEL conference was held at the Eugenides Foundation, Athens on the 4–6 November 2018. This is the report from that meeting.
To Set the Scene
Klara Capova (University of Durham) pointed out that there are only a few human questions as fundamental as “how did all this life around us come into existence?” and “are we alone in the Universe?” In order to answer these questions, I reflect on this speaker’s words, at the NoRCEL meeting in November 2018, when she stated that: “astrobiology is a multidisciplinary field of science encompassing life sciences (i.e., biology, chemistry and physics) and planetary as well as the social sciences and humanities. Such complexity poses a challenge of how to effectively communicate and disseminate this wide and innovative field of knowledge to audiences both within and outside academia [1]. In addition, she adds that scientists today take a proactive role in airing and sharing within scientific communities as well as engaging and enthusing the general public. This should pave the way forward for the better understanding of the field of astrobiology.
The Origin of Life?
Broadly there are three front runners when it comes to the origin of life. The metabolism first hypothesis promulgates that pathways, cycles and hypercycles came first; the genetic first hypothesis claims that a chemical informational system was the first to emerge. This is simply because both metabolism and genetics are interlinked in that, as observed in contemporary life, proteins (being part and parcel of metabolism) are needed to carry out the necessary metabolic reactions to make the information carrying molecules. Genetics, or strictly speaking, the chemical informational systems, are needed to instruct the formation of proteins; in short, an endless chicken and egg situation. The third hypothesis is vesicle first with its assertion being that life’s reactants need to be in the vicinity of one another in order for their chemical reactions to take place; reactions in an open body of water will not occur because reactants will probably not “bump” into one another easily due to the dilution effect of water. In any case, two of life’s initial challenges would be (a) that every condensation (of monomers) reaction requires a huge amount of a typically specialised form of energy (e.g., ATP) because the dimer formed is at a higher level compared to the two monomers from which it is made, on the energy diagram; that is to say the reactions are endothermic and (b) every condensation reaction generates a water molecule; since life emerged, it is believed, in watery environments, this means that condensation reactions would be diluting the reactant with the continued process of polymerisation and so Nolan Grunska (Montana State University) posed a question: “Are scientific models of life testable?” He used Simpson’s Paradox to expound the answer. The Paradox is a statistical modelling whereby the observed probabilistic “effects” in individual data sets could be considerably different from when the individual data groups are combined into a single set. Such probabilistic effects may either disappear or reverse trends observed in the individual data sets. Simpson’s Paradox was applied to both metabolism and genetic hypotheses. Grunska argued that even though the relevant chemical reactions involved in the emergence of life might have been deficient in many of the hypothesized settings—as they have been shown to be in various lab experiments—when reactions of life are taken or considered collectively they could be significant; a reversal characteristic of Simpson’s Paradox might show up, and Grunska provided examples of hypothetical data sets in the context of each emergence of life hypotheses where a Simpson’s Paradox occurred. In these data sets, ratios of acetyl-CoA [metabolism] and ribose [genetic], to non-functional molecules in the proposed chemical reactions, were maintained from those found in relevant lab experiments used to argue that either theory is implausible due to chemical inefficiency. Thus, the Paradox helps to resolve differences in either of the theories on the grounds that the initial reactions necessitated by them are chemically inefficient, and thus could not lead to the emergence of life, showing instead that they may each still be plausible. As for testability, a scientific model is testable in two senses, either in a weak sense or in a strong sense. The model-based conjecture that life could emerge in either of the modes of the hypotheses, metabolic first or the genetic first, is testable in the weak sense because the model Grunska proposed is logically consistent and does not violate laws of physico-chemistry. However, whether the model can be regarded as testable in the strong sense remains a desideratum for further investigation [2,3].
Metabolism First
Frances Westall (CNRS-Centre de Biophysique Moléculaire) spoke about the metabolism first hypothesis and the possible site(s) at which life may have emerged. She proposed that the sedimentary layer between oceanic crusts and seawater, flushed by hydrothermal fluids, was the site for the origin of life, (as exemplified by rocks from the 3.5–3.33 billion-year old Barberton Greenstone Belt, South Africa), as opposed to the more popular alkaline hydrothermal vents hypothesis. Sedimentary rocks from the Barberton Greenstone Belt site suggest that the early sediments from the Hadean (~4.4–4.0 billion-year) were likely characterised by physical and chemical gradients, such as ionic concentrations, pH, and temperature gradients; this set-up being akin to miniature chemical reactors for the production and complexification of prebiotic molecules which led to the emergence of life—Figure 1 [4].
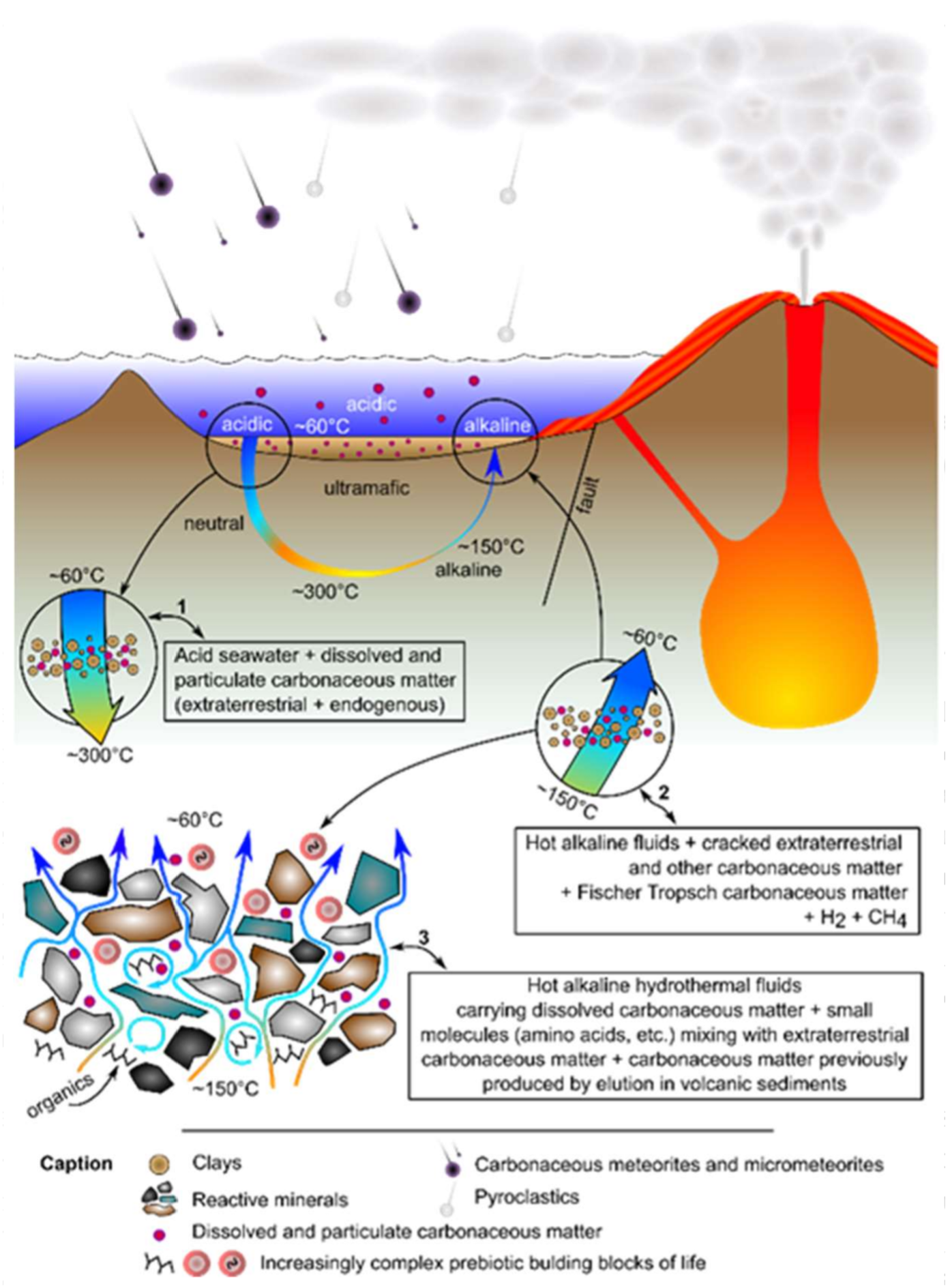
Figure 1. Schematic synthesis of the proposed Hadean, hydrothermal-sedimentary micro-reactor environment for the complexification of prebiotic chemistry.
Vladimir Kompanichenko (Russian Academy of Science, Birobidzhan), proposed a mechanism by which life could have emerged entitled: “Thermodynamic inversion: origin of living systems.” The primary living units (called “probionts”) cannot emerge out of organic compounds simply by continuous chemical complication of prebiotic macromolecules and microsystems. The complication must be accompanied by the radical thermodynamic transformation of prebiotic microsystems that resulted in the acquired ability to extract free energy from the environment and export entropy. He called this transformation “thermodynamic inversion” [5]. The inversion may occur by means of the efficient (intensified) response of the microsystems on the oscillations of physico-chemical parameters in hydrothermal environments. In this case the surplus available free energy within a microsystem, when combined with the informational modality, facilitates its conversion into a new microsystem—a living probiont. Figure 2 is a schematic representation of an oscillating prebiotic microsystem whereby the primary living unit (probiont) emerges as a result of the surplus free energy (prevalent over entropy) as shown by the red arrows. To verify his own concept, Kompanichenko has put forward a new experimental set-up pertaining to prebiotic chemistry under oscillating conditions [6].
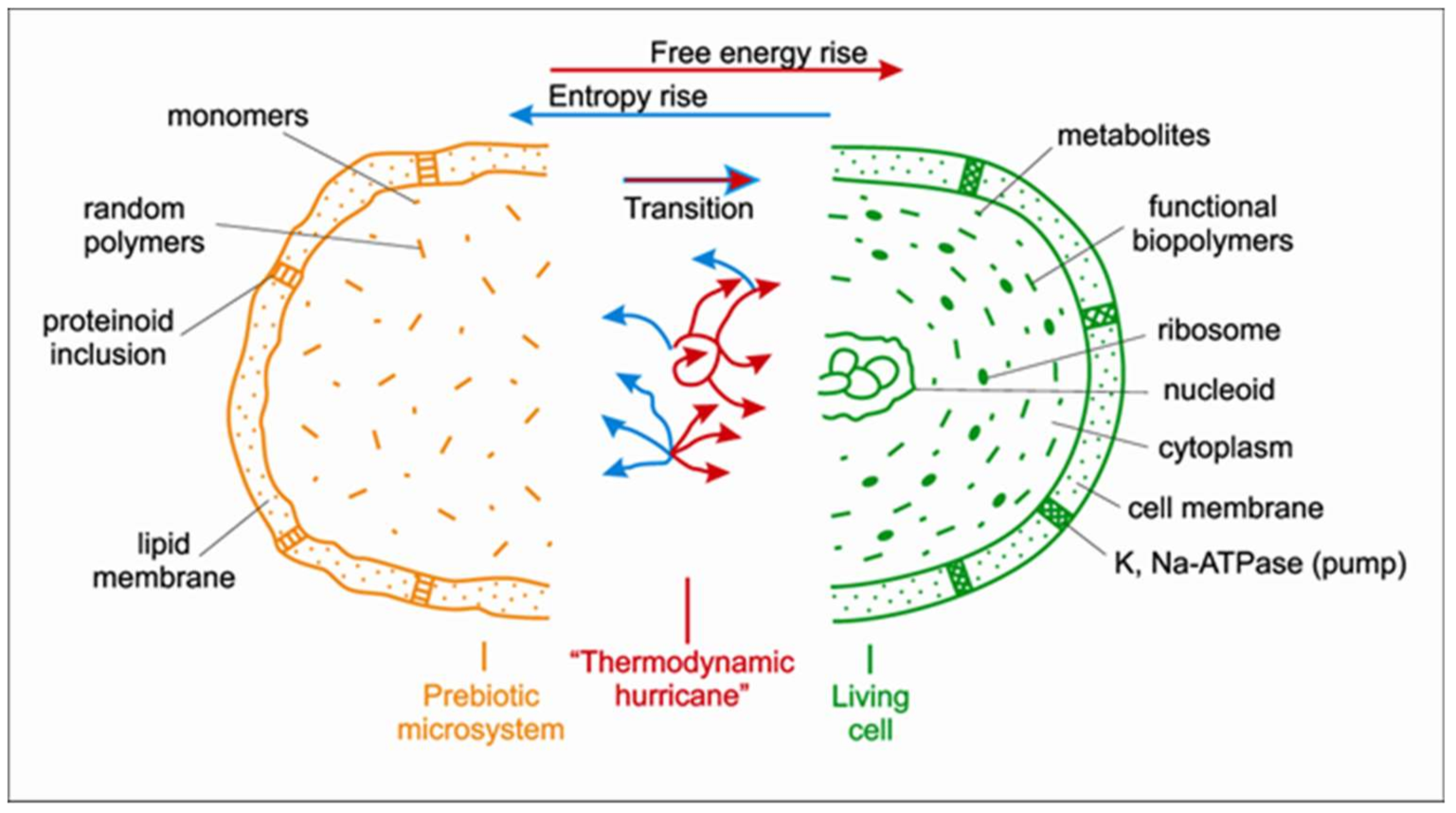
Figure 2. The left hand, orange half comprises the prebiotic microsystem being composed of a random mix of monomers and other relevant smaller, simpler molecules, as well as polymers. The central part represents the transition of the prebiotic microsystem into simplest living entity (initial cell), when the processes producing free energy (red arrows) become more efficient than ones producing entropy (turquoise arrows); this stage is conditionally termed: “thermodynamic hurricane”. The right-hand green half shows the emergence of the simplest living entity, namely probiont, whose further evolution led to formation of the main cellular structures such as nucleoid with its circular DNA, ribosomes and cell membrane. (Readers may wish to consult Kompanichenko, 2017, book entitled: “Thermodynamic Inversion: Origin of Living Systems”).
Rowena Ball (Australian National University) outlined her radical hypothesis based on well-known physico-chemical chemistry entitled: “the HP crucible hypothesis”. This holds that hydrogen peroxide (HP) may have played an instrumental role in the origin of life and evolution of the first living systems. There are three main strands to the hypothesis, namely that (a) as an oxidation of the thiosulfate/hydrogen peroxide (THP) redox oscillator, thereby providing a periodic power source; (b) HP may act as an agent of molecular change and evolution, and mediator of homochirality; and (c) the THP oscillator is subject to Brownian input perturbations and is life-enabling, as it produces a weighted distribution of output of thermal fluctuations that favours polymerisation and chemical diversification over chemical degradation and simplification. The HP crucible hypothesis can help to clarify the hero and villain roles of HP in cell function and can explain the singularity of life: of necessity, life evolved an armoury of catalases early on, the continuing and all-pervasive presence of which prevents HP from accumulating anywhere in sufficient quantities to host a second origin [7–9]. Helen Hansma (University of California at Santa Barbara) posited that life might have started between sheets of mica, powered by the mechanical energy (mechanochemistry) of moving mica sheets. The idea being that mica has sufficient space between the sheets for life’s precursor molecules to be concentrated and that mica sheets have high concentrations of potassium ions (K+), like the protoplasm of all cells. The black mica Biotite might be the best mica for life’s origins because it is high in the iron needed for redox chemistry [10–13].
Genetics First
Metabolism and genetics are interlinked because at the heart of the metabolism first hypothesis lay ultra-efficient and ultra-specific enzymes which carry out all the chemical reactions within a cell. These enzymes are made under specific instructions from chemical informational systems and the specific chemicals from which the informational systems are made are types of polymers, specifically polyribonucleotides (RNAs); these polymers are specifically composed from nucleotides of guanine (G), cytosine (C), adenine and uracil (U). The essence of these RNAs is that they have several attributes, namely they can act as a repository for rudimentary chemical information (cf tRNA, mRNA and RNA viruses); carry out self-replication via template directed propagation; self-process, e.g., carry out activities displayed by group1 introns [14]; suppress gene expression via RNA interference (RNAi) [15]; and act as RNA based catalysts, namely ribozymes [16,17]. RNAs can also form 4 levels of structure as follows: primary, e.g., small sequence motif; secondary, e.g., 5S, rRNA motifs; tertiary as seen in leadzymes, hammerhead and twister ribozymes, as well as tRNA; and finally, quaternary structures, the prime example being ribosomes. Since RNAs broadly only use four types of bases to achieve all four levels of structural integrity, this means that an RNA is a versatile molecule with enormous possibilities. Three of these possibilities were highlighted by Sohan Jheeta (NoR CEL, UK), declaring that structural integrity defines the function and range of activities that an RNA can display, i.e., self-organisation, self-assembly and instructed chemistry. The self-organisation aided in the formation of pathways, cycles and hypercycles, which led to the eventual formation of the super hypercomplex chemistry which we call life. The self-assemblies are systems that tend to gather themselves from their individual parts, forming 3D nano-structures, whilst at the same time minimising the free energy within the final attained structure, a prime example of self-assembly would be viral capsid formation from capsomeres (monomers); RNAs plus peptides leads to the assemblage of ribonucleoproteins structures, miniature organic nano-machines and the ultimate such machine is a self-assembly of ribosomes from small and large sub-units of ribonucleoproteins. At the pinnacle of structural integrity lies an instructed chemistry. In essence this chemistry is a cooperation between three types of RNAs namely tRNA, mRNA and rRNA. This cooperation is well documented and so it will not be reappraised here fully. In short, both small and large subunits of rRNA attach to the start codon (AUG) on an mRNA which carries specific chemical informational instructions from the DNA (ie a transcript of DNA with specific base sequence). The ribosome is now ready for protein synthesis; the synthesis begins when tRNA with UAC anticodon attaches to the AUG codon at the head of mRNA. The order of stitching together of amino acid is dictated by the triplicate base sequence (i.e., codons) on the mRNA. The amino acids are stitched together at the “P” site located within the ribosome when an activated amino acid, carried by the tRNA (aa~tRNA), is brought to the site with the eventual formation of a peptide bond. The important part during the synthesis of protein is the crosstalk between the aa~tRNA and mRNA, as this “interaction” between two RNA molecules may have evolved during the self-organisation era mentioned previously [18], whereas the instructed chemistry came both after self-organisation and self-assembly. All three mechanisms are necessary for the emergence of life. Jheeta refers to this as “probable chemistry” because the chemistry from which life emerged was more likely to happen than not—i.e., more probable than improbable [19,20]. Sandeep Ameta (ESPCI Paris) asserted that life originated as a result of networks of autocatalytic catalysts, based on RNA (ie ribozymes) which gradually evolved during the RNA world epoch. The premise being that RNA molecules can act as a repository of information as well as being catalysts, albeit slowly; in the contemporary cell these functions are carried out by DNA and proteins respectively. His studies involve droplet-based microfluidics and single droplet-level sequencing on RNA fragments derived from group I intron of Azoarcus bacterium. This study highlighted that rudimentary RNA-based networks possess properties critical to Darwinian evolution. He further showed that these properties are controlled by network structures and there are trade-offs between them. The study experimentally demonstrate that chemical systems based on autocatalytic networks are indeed evolvable [21–23]. But how were the monomers of RNA made in the first place? When engaging in discussions on prebiotic chemistry, questions which often crop up are as follows: how were simpler prebiotic precursor molecules (e.g., formamide) converted into the necessary life friendly organic molecules? That is, how were the necessary basic chemical informational molecules made other than being delivered onto the Earth by extra-terrestrial impactors? The chemical informational molecules, in this respect, being purine and pyrimidine nucleic acid bases; the four main ones are cytosine, guanine, adenine and uracil. Asif Iqubal (Indian Institute of Technology Roorkee) addresses these particular questions. He floats the idea that such nitrogenous bases of purine and pyrimidine can be made on Earth, on the surface of iron oxide nano-particles, namely NiFe2O4, CoFe2O4, CuFe2O4, ZnFe2O4 and MnFe2O4 as these divalent metal ions in iron oxide make them excellent catalytic systems. These minerals may have been present on Earth by 300 million years after its formation [4,24,25]. Konstantinos Papasakellariou (Agricultural University of Athens) delivered an impressive oral presentation on a high throughput computational approach for the fractionation of RNA species from whole transcriptomes. ncRNA-based interactions have a well-characterised genetic impact inside cells. He presented preliminary results on quantifying the naturally occurring RNA diversity, using a combination of previously developed algorithms, and RNA-seq data derived from photosynthetic organisms. The aim was to map the primary pools of RNA sequences from different sub-cellular loci, towards extrapolation of specific RNA classes and then to group such biomolecules under distinct categories of similarity with respect to their size, sequence, biophysical type and compartmental origin. These findings were discussed in relation to the evolutionary mobility of potent RNA signals among organellar & nuclear genomes [12,26,27]. The implication of this research is that ncRNA molecules with a particular structural configuration and respective functional attributes can encourage biodiversity since such structural and functional aspects of ncRNA also affect redox and genetic characters leading to co-evolution.
Vesicles First
Next, Doron Lancet (Weizmann Institute of Science, Israel) disseminated the idea of spontaneous self-assembly of compositional lipid catalytic networks. This concept is not as implausible as it may seem, in that there are molecules in nature that can self-assemble; a typical example being heterogeneous molecules that have both hydrophobic tail and hydrophilic headgroups—i.e., amphiphilic molecules. These amphiphiles readily and spontaneously self-assemble to form micelles and vesicles. Such assemblies are necessary to bring life’s molecules together within the vicinity of one another, without the diluting effect of an open body of water, in order that they can react. Lancet added another dimension to the idea of self-assembly via proposing a rigorous chemical kinetics simulated model termed: Graded Autocatalysis Replication Domain (GARD). Lancet’s kinetic simulations show that specific GARD lipid assemblies (“composomes”) may undergo compositional reproduction through mutual catalysis, making them candidates for early replicators, prior to the emergence of RNA, giving credence to the vesicles first hypothesis. He also shows through a variant kinetic model, Metabolic GARD, that lipids may gradually “grow” short segments of sequence-based informational biopolymers as part of their polar headgroup, a first step towards the appearance of proteins and RNA. The thrust of his argument is thus that these units pre-dated the traditionally hypothesized chemical information-first polymers, namely RNA and DNA. Being capable of reproduction with mutations, M-GARD’s composomes may have ultimately evolved to become protocells. Lancet estimates this may have taken place several hundreds of millions of years after the Earth cooled down, to reach the Last Universal Common Ancestor (LUCA) at just prior to 3.5 billion years ago. The latter time frame being the point at which the existence of fossils pertaining to the unicellular life forms have been confirmed with a degree of certainty [28–31].
Gene Transfer
Generally, gene transfer is referred to as horizontal gene transfer and historically there are three major types of mechanism by which mobile genetic elements are transferred between organisms. These mechanisms are as follows: transformation, conjugation and transduction. The evolution of bioenergetic variety within prokaryotes is characterised by a wide spectrum of metabolic diversities. This spectrum of diversity allowed prokaryotes to occupy various habitats including extreme environments. Other than oxygenic photosynthesis and oxidative phosphorylation, prokaryotes utilise various chemo or lithotrophic strategies which include anoxygenic photosynthesis, both iron oxidation and reduction, sulphate reduction, and methanogenesis. The bioenergetic pathways have been studied extensively in vitro in detail but not much about the emergence of these pathways and their evolutionary relationships is known. In order to elucidate the emergence and evolutionary relationships, Vassiliki Lila Koumandou (Agricultural University of Athens) mapped the distribution of nine bioenergetic modes on a phylogenetic tree based on 16S rRNA sequences from 272 species of fully sequenced prokaryotes, which represented the full diversity of prokaryotic ancestries. Her studies highlighted the irregular distribution of many pathways across different lineages which brought into focus that either as many as 26 independent origins, or 17 horizontal gene transfer events had occurred [32]. Further, comparative genomics and phylogenetic analysis of all subunits of the F0F1 ATP synthase, common to most bacterial lineages, revealed an ancient origin of this protein complex, and no clustering based on a bioenergetic mode. This suggested that no special modifications were needed for the ATP synthase to function with different electron transport chains [32]. She also reported that the distribution and evolutionary relationships of different b-type cytochromes within the electron transport chains were investigated and the results showed that an ancient origin and diversification of cytochromes had occurred [33] and that these cytochromes are not specific to any one pathway in particular. Although Koumandou expounded the possible effect of these bioenergetic pathways on the dynamics of the human gut microbial community [34], in the context of origin of life her results demonstrated (a) irregular distribution of pathways among different lineages due to gene transfer events; (b) ancient origin of the F0F1 ATP synthase and that no special modifications was necessary; and (c) an ancient origin and diversification of b-type cytochromes.
In recent times, two additional gene transfer systems have been identified, namely gene transfer agents (GTAs) and the extracellular vehicles (EVs); the former is akin to transduction and the latter is similar to exocytosis and is prevalent in most organisms within the three domains of life, namely Archaea, Bacteria and Eukarya. Andrew Lang (Memorial University of Newfoundland, Canada) promulgated that GTAs are bacteriophage-like particles produced by some prokaryotes that exclusively package small fragments of cellular DNA. Production of GTA particles by the alphaproteobacterium Rhodobacter capsulatus occurs in a small subset of the population (<3%), with these cells lysing to release particles that can then transfer the packaged DNA to other cells in the population. The production of GTAs in R. capsulatus is controlled by several regulatory systems and coordinated with the capability of non-producing cells to become competent to receive DNA from the GTA particles [31,35,36]. Sherri Christian (Memorial University of Newfoundland, Canada) disseminated that EVs are heterogeneous groups of lipid-encapsulated nano-particles that can be secreted from the cytoplasm or bud from cellular membranes. EVs can be produced by any organism, from prokaryotes to eukaryotes, and are found in all bodily fluids in multicellular organisms. EVs contain bioactive RNA, DNA and protein that can affect the function of recipient cells. However, the role of EVs in cell-cell communication in normal development or disease states is not fully understood. She has recently found that membrane-derived EVs are released from B-lymphocytes in response to stimulation of a cell surface receptor, CD24. Evidence suggests that these EVs can alter the microenvironment to affect cellular development. [31,37,38]. What relevance, if any, does Lang and Christian’s research bear to the origin of life? One cannot help wondering whether such mechanisms were both relevant and present at the earliest stages, perhaps during LUCA epoch, of life’s emergence.
Virus System
In an historical setting, viruses are primarily defined as obligate intercellular parasites: this much is true of all viruses. They are parasites because they lack a variety of defined semi-permeable boundaries such as cell membranes as well as containing internal large-scale structures (mitochondria and ribosomes) depending on the class of cells they belong to; and they also lack cellular functional and vibrant biochemistry. Viruses typically contain only one type of nucleic acid, either RNA or DNA, wrapped in an outer casing, namely capsids. Tamir Tuller (Tel Aviv University) added that the inanimate informational chemical systems, namely the RNA and the DNA modalities, are responsible for all intracellular processes within all life forms and are also connected to the phenotypes of organisms. The same informational chemical system is also true of viruses, although the intracellular processes within viruses is absent. It is sufficient to say that once the viral informational chemical systems are within living cellular life forms, the “effect” is akin to those mechanistic processes that occur in cellular cells, with the major difference being that the “parts being formed” are of viral nature. Despite this obvious difference between cellular and viral entities, it is the expression of informational chemical systems (i.e., gene expression) that connects the inanimate genetic elements to the chemical processes and the phenotypes. In his oral presentation Tuller demonstrated the computational models that could predict various aspects of the biophysics of gene expression and evaluated the understanding of the evolution of cellular organisms and viruses further—further information on this interesting topic can be found in his papers [39–41].
Bio-Signals
How can life be determined in the wider Universe? There are primarily two ways to achieve the feat of ascertaining whether or not life could exist elsewhere in the Universe, namely remote sensing or in situ detection of bio-signals. Remote sensing involves detecting organic chemical signals emanating from distant dark molecular clouds, circumstellar disks (e.g., PDS 70 as found in the constellation Centaurus) and exoplanetary atmospheres and can be confirmed by an appropriately tuned antenna of a radio-telescope, such as the one at Jodrell Bank in the UK or space-based telescopes (e.g., the Hubble Space Telescope). Chemical signals emanate due to the fact that molecules vibrate when electromagnetic radiation impinges on them. When a molecule absorbs electromagnetic radiation, its bonds expand. Upon bond relaxation the molecule emits infrared radiation between 4000–400 cm−1 wavenumber range. For example, carbon dioxide (CO2) can be detected at the following wavenumbers: 660, 830, 1399, 1546, 2341 cm−1 [42,43]. For further information the reader may be interested in reading the following entitled paper: “Final frontiers: the hunt for life elsewhere in the Universe” [44]. These signals could be detected by a radio-telescopes. Detection of chemical signals from organic molecules (and the presence of extant life if present) could be determined by in situ instrumentation, such as Time of Flight (ToF) mass spectrometry or dynamic mass spectrometry, either based on ion sources using sputtering methods, or on laser desorption using high energy pulsed lasers. Elias Chatzitheodoridis (National Technical University of Athens) gave us an insight into these state-of-the-art instruments and the methods for detecting isolated organic molecules from rocks using the spectrometry algorithm mentioned above. He presented results from the deployment of such instrumentation on terrestrial rocks; carbonaceous and Martian meteorites; and in situ deployment of COSIMA (COmetary Secondary Ion Mass Analyzer) on the surface of comet 67P/Churyumov–Gerasimenko during the ESA’s ROSETTA mission, Launch date: 2 March 2004 [45,46] showing that detecting in situ chemical signals is a realistic possibility and highly recommended. Stefan Fox (University of Hohenheim) delivered an oral presentation on searching for signs of life elsewhere in our Solar System and on exoplanets further afield. He pointed out that at times, false positive results are generated during such remote searches. These false positive bio-signals may be of abiotic nature—for example, although porphyrins, in particular metalloporphyrins, are often thought to be of biological origin, the very same molecules could also be made abiotically [31,47,48], thus giving the impression of the presence of tentative cellular life forms. Since there are many such molecules (e.g., methane and see Kotsyurbenko below) that have an abiotic origin, these cannot be excluded, meaning that the risk of misinterpretations is continually ever-present [49]. Therefore, sophisticated combinations of analytical methods are necessary. From a technical point of view, laboratories, in general, aim to produce the finest tests and methodologies with the good intention of generating viable best results, but the truth of the matter is that there is still a high risk of terrestrial contaminations, which, in essence, makes a good case for a very expensive sample return mission. However, possible bio-signals (e.g., methane) could also be determined by in situ instrumentation from local solar bodies (e.g., Mars and Titan, Saturn’s moon). In addition to standard methods, e.g., chromatographic and spectroscopic methods, high resolution and high accuracy mass spectrometry generate the necessary isotopic ratios; such isotopic ratios of hydrogen, nitrogen and carbon in molecules will throw some light on the elucidation of their formation in space in general. He points out that both remote and in situ detection of bio-signals is problematic because the Universe is colossal and so the distances involved are immense. This means with increasing distances the [background] noise aspect of signal-to-noise ratio makes received signals indecipherable. Oleg Kotsyurbenko (Yugra State University, Russia) gives us some insights into the origin of life via asserting that methanogenic microbial communities are complex “trophically” interconnected within biological niches. The way to unlock the interaction between these communities is to follow the energy trail within a particular group, noting that the hardy members of methanogens are the end-of-the-line microbes which flourish depending on substrates provided by other microorganisms in the trophic chain. Evolutionarily, the mode of energy and carbon obtained by methanogens makes them the most ancient microbes on Earth especially compared to those microbes belonging to the domain: Bacteria. Methanogens belong to the domain: Archaea. Kotsyurbenko highlighted that methanogens produce methane and a sustained production of this gas, as compared to abiotically produced methane, would be a clear indication of the presence of life on exoplanets; this would be particularly relevant when other bio-signals (e.g., the presence of water and ozone etc. [44] emanating from exoplanet’s surface are taken into consideration [50,51]. Christos Georgiou (University of Patras, Greece) extrapolated the possibility of detecting bio-signals from Martian life; and on Jupiter’s moon, Europa, as well as on Saturn’s moon, Enceladus using the functional properties of terrestrial lipids and amino acids; and catalytic and structural aspects of enzymes. Georgiou’s oral presentation was in two parts as follows: (a) amphiphilic hydrocarbons with 4C to 9C chains as found in meteorites will not do because they do not form stable, fluid lipid bilayers; in comparation 16C or longer chains, the type that are found in terrestrial membranes, are ideal for bio-signals of extra-terrestrial life, especially if these transmembrane chains are even-numbered. In addition, (b) certain patterns which are common to certain organic molecules, in particular amino acids, that are universally associated with biological functions give powerful clues to life in the Solar System: the premise for the second part is that some hydrolytic catalytic groups such as imidazole (in His), thiol (in Cys), guanidinium (in Arg), and amide (in Asn, Gln) are found in the enzymes of contemporary living entities, especially those enzymes that possess exceptionally high throughput. More importantly, these catalytic groups do not feature in meteorites, which means that these groups can be used as indicators of extra-terrestrial life forms upon identification by existing methodologies—for example, ultrasensitive fluorescent assays, which uses certain artificial hydrolytic substrates, namely umbelliferone and 6-methoxynaphthaldehyde. Using such techniques, further confirmation can be obtained from these amino acids as they have a high propensity to form α-helical intra-membrane peptide domains and therefore may serve as early transmembrane transporters in protocells, as well as catalysing simple biochemical reactions. Furthermore, the chemical groups mentioned above also have high affinity to form 3D-structures in terrestrial catalysts such as those found in extremophiles including thermophiles, psychrophiles and halophiles; likewise these amino acids may play a significant biochemical role in extra-terrestrial life forms found on the solar bodies within our Solar System and even further afield elsewhere in the Universe [52,53].
Space—The Final Frontiers
Pauli E Laine (University of Jyväskylä, Finland) expounded on the two interlinked questions as follows: does life always emerge under the right conditions and is life on Earth a unique phenomenon in the observable Universe? Moreover, he hypothesised that analogues in modern cosmology have counterparts in life, pointing out that life is a phenomenon that emerges from a micro-scale physiochemical landscape. Once established, it undergoes inflationary expansion in the form of speciation and bio-diversity, just as the Universe inflated due to quantum fluctuation to large-scale galaxies, star clusters and solar systems. Furthermore, he argued that the likelihood of the phenomenon will actually increase the possibility of extra-terrestrial life existing in the Universe [54,55]. On the question of solar systems, Taichi Uyama (University of Tokyo) is working on direct imaging methods and the main targets of his research are young stellar objects (YSOs). Such objects are less than 10 million years old and typically have protoplanetary disks. These disks are places where planets are formed and thus eventually, solar systems are accreted. It is these that Uyama is interested in imaging and thus a better understanding in space exploration and the potential for life in the Universe looks promising [56–58]. Andjelka Kovacˇevic´ (University of Belgrade, Serbia) concentrated on a novel study on peculiar radiation effects of nearby (z < 0.5) supermassive black holes (SMBH) on the erosion of exoplanetary atmospheres and compared it with the Milky Way’s SMBH. This peculiarity is detected via statistical data analysis of 54 exoplanets—16 Sagittarius Window Eclipsing Extrasolar Planet Search (SWEEPS) planets and 38 Earth-like exoplanets—and 33,350 SMBHs at z < 0.5. All SWEEPS planets are hot Jupiters—i.e., Jupiter-like planets with very small semimajor axes of their orbits and they are located in the vicinity (or bulge) of our galactic SMBH, named Sgr A*, at distances of approximately 760 pc. This study shows that atmospheric erosion of the SWEEPS planets, due to irradiation by an active Sgr A*, results in 10 SWEEPS losing mass greater than that of Earth’s atmosphere, and 13 of the exoplanets would have lost mass greater than the current atmosphere of Mars. This happened in the 50 Myrs of an active galactic nucleus (AGN) lifetime of Sgr A*. Earth-like planets could lose even 1% of their atmospheres purely because of radiation from close by SMBHs outside of the Milky Way. Since atmospheric mass loss is planet density dependent, one can expect a higher mass loss percentage for Jovian planets having lower densities [59]. Based on this analysis, one of the beneficial SMBH radiation effects in our Galaxy could be stripping unhabitable giant gaseous planets of thick hydrogen-helium atmospheres, and converting them into rocky super-Earths [60]. However, taking into account that Jupiter-like planets contain massive atmospheres, this effect would be highly dependent on a Jupiter-like planet’s evolutionary stage, amongst other numerous physical factors [61]. Claudio Maccone (Istituto Nazionale di Astrofisica, Italy) gave ontogenesis a mathematical treatment as might be applicable to life elsewhere in the Universe; ontogenesis is a developmental phase of an organism during the time frame from birth to puberty. During this period an organism uses the maximum amount of energy (compared to any other point during its life) and this can be measured as the total energy (Joules, J) used by an organism during ontogenesis. The crux of the ontogenesis phase is that an organism is mature enough to reproduce the next generation of their species-kind. Whilst the ontogenesis phase may be applicable to all living entities on Earth, in respect of his oral presentation he was primarily concerned with human-kind. Thus, in mathematical terms Maccone describes the ontogenesis phase as the “logpar” power curve as described in his theory entitled: “Evo-SETI Theory” [62,63]. His latest results on the total energy of ontogenesis are as follows: (a) the energy of ontogenesis could be tweaked by reducing the error functions typically used in probability theory; and (b) the ontogenesis phase uses the highest amount of energy and it reduces when an organism becomes adult, a time of intellectual maturity, and makes a decision to reproduce. The conclusion being that intelligent life forms elsewhere in the Universe might follow similar trends as posited by Evo-SETI Theory. Martin Dominik (University of St Andrews) argued that a key reason why attempts towards a universal definition of “life” have failed is that at least two very different concepts are being conflated and would be better if separated. He suggested distinguishing “eLife” (“e” as in “evolutionary”) to refer to life as a collective process, from “iLife” (“i” as in “individual”) to refer to life as a property of individual beings. We can only claim a detection of “life” once it is clear what it means, and it might be sensible to adopt definitions that do not match the historic understanding of “life”; what the exact historic understanding of life is, he did not expound further. My feeling is that he means when comparing inconsistencies and deficiencies of both inanimate replicators (e.g., fire) with the biological ones, including their peculiarities, as in mules are sterile and are therefore by definition, non-living. In contrast, all discussions about “habitability” fall short of providing crucial links required to further foster our understanding. He concluded that exploring the Universe in the search for ourselves is a venture that is both most fascinating and most frustrating at the same time.
What’s Next?
The contemporary biochemical reactions of life in cellular biology teach us that these reactions occur in a series of steps, one after another and another and so on; such a series of reactions can form a pathway which may perhaps link to another pathway, and then form a cycle; the cycles and pathways then evolve into hypercycles—a concept of chemical evolution has thus emerged and life followed soon afterwards, within a relatively short time frame of 300 million years [64]. More importantly, these series of steps are followed faithfully without side-stepping or by-passing any reactions, at appropriate and constant rates and generally at a well-maintained pH, salinity, osmotic pressure and temperature environments etc. The proof of this sequential chemistry is only betrayed by the fact that viruses completely lack gradual stepwise chemistry; this defines the difference between viruses and cellular life forms, noting that viruses were present at the time of LUCA [31,65,66].
The same gradual stepwise chemistry, has not altered since the era of prebiotic chemistry, meaning that there must have been probable reactions which occurred step by step; a chemical behaviour which is counter-intuitive because nature is raw and, at least in theory, chemical reactions which could proceed in any infinitesimal direction in a chaotic fashion, yet chemical reactions hurtled towards chemical evolution and the eventual emergence of life. If one is to examine any contemporary cellular life form, one would discover that the cell is packed full of chemical reaction by-products (e.g., CO2) and newly made products (proteins), as well as activated reactants (e.g., 20 different types of charged and neutral amino acids and nucleotides). Also present, depending on the domain of life, are ribosomes, endoplasmic reticula, lysosomes, vacuoles, a full complement of DNA, a raft of different RNAs, plasmids, mitochondria, centrioles, Golgi apparatus, starch granules, protein active sites, proteolytic and nucleolytic enzymes and nucleases, as well as electrolytes – including ions of hydrogen (H+), potassium (K+), sodium (Na+), chlorine (Cl−), as well as amphoteric and triglyceride molecules. If molecules within a cell have to travel from the ribosome, where they are synthesised, to the place where they are needed, this is a herculean task because the molecules will be blindly knocking into one another, rebounding and also being attacked by various radicals and proteolytic enzymes; its “safe” passage will be blocked by other larger molecules and nano-machines (e.g., introns), not forgetting that water molecules, being bipolar, will be a major barrier too. In addition, a newly made protein will probably also be in danger of being attacked by proteolytic enzymes. On the molecule’s arrival at its place of destiny in the cell, the active site on the recipient molecule may be pointing away from the incoming molecule; the correct orientation of both molecules has to be synchronised before any discernible intended activity can take place. However, the cell still manages to carry on with the business of living, performing appropriately necessary reactions and replications often on nanoscale time-frames. All the while the cell’s environment is chaotic rather than a cosy organised place—is this an example of the probable chemistry that was prevalent during the prebiotic epoch? It should be remembered that life demands an unchanging landscape and so the cell’s chaotic environment is a reflection of that bygone era. In trying to fully explain the chemical evolution of life, unfortunately we only have conjecture at this stage – this report highlights that very point, as all the scientists featured have outlined a theoretical frame-work, with very little concrete chemistry. Let me explain further, any experimental prebiotic chemistry which is regularly carried out in astrobiology labs the world over appears to be a somewhat “school bench” type of chemistry. That is to say, these are generally a single reaction at a time, which generates a particular product(s)—“C” in Equation (1). This product is then placed in another test tube in order to carry out the next reaction (Equation (2)) and so on.
TEST TUBE 1: A + B →C + D (1)
TEST TUBE 2: C + E →F + G (2)
It is similar to adding acid to an alkaline to make salt. This salt is then used to carry out another follow up experiment—see “prebiotic RNA synthesis by montmorillonite catalysis” [67]. There is a “but” here and it is a big but, as there is no purification and concentration of reactants taking place in nature; there are no measured moles and defined temperatures and pressures; there is no washing of clay or preparing of this or that crystal. Nature is as it is: raw, and because it had a chemistry taking place about which we are still in the dark, what is needed is a new approach and new way of thinking and appreciation of the concept of “PROBABLE CHEMISTRY”.
Acknowledgments: The author would like to offer his thanks to all the contributors to this paper for double checking their content.
References
- Capova, K.A.; Persson, E.; Milligan, T.; Dunér, D. Society, Worldview and Outreach. In Astrobiology and Society in Europe Today; Springer: Cambridge, UK, 2018; pp. 19–24.
- Bandyopadhyay, P.S.; Raghavan, R.V.; Dcruz, D.W.; Brittan, G. Truths about Simpson’s Paradox: Saving the paradox from falsity. In ICLA 2015: Logic and Its Applications; Lecture Notes in Computer Science; Banerjee, M., Krishna, S.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 8923.
- Bandyopadhyay, P.S.; Beard, T.E.; Greenwood, M.C.; Bertasso, M.P.; Peters, J.W. Why Need a Model? The Debate over the Origin of Life Theories and a Lesson from Simpson’s Paradox. In Proceedings of the Epistemology of Modeling and Simulation Conference, Pittsburgh, PA, USA, 1–3 April 2011; University of Pittsburgh: Pittsburgh, PA, USA.
- Westall, F.; Hickman-Lewis, K.; Hinman, N.; Gautret, P.; Campbell, K.A.; Bréhéret, J.G.; Foucher, F.; Hubert, A.; Sorieul, S.; Dass, A.V.; et al. A Hydrothermal-Sedimentary Context for the Origin of Life. Astrobiology 2018, 18, 259–293. [CrossRef]
- Kompanichenko, V.N. Thermodynamic Inversion: Origin of Living Systems; Springer: Cham, Switzerland, 2017; 275p.
- Kompanichenko, V. The Rise of a Habitable Planet: Four Required Conditions for the Origin of Life in the Universe. Geosciences 2019, 9, 92. [CrossRef]
- Ball, R.; Brindley, J. The Power Without the Glory: Multiple Roles of Hydrogen Peroxide in Mediating the Origin of Life. Astrobiology 2019, 19, 675–684. [CrossRef]
- Ball, R.; Brindley, J. Toy trains, loaded dice and the origin of life: dimerization on mineral surfaces under periodic drive with Gaussian inputs. Soc. Open Sci. 2017, 4, 170141. [CrossRef]
- Ball, R.; Brindley, J. The Life Story of Hydrogen Peroxide III: Chirality and Physical Effects at the Dawn of Life. Life Evol. Biosph. 2016, 46, 81–93. [CrossRef]
- Hansma, H.G. Better than Membranes at the Origin of Life? Life 2017, 7, 28. [CrossRef]
- Hansma, H.G. The Power of Crowding for the Origins of Life. Life Evol. Biosph. 2014, 44, 307–311. [CrossRef]
- Jheeta, S. The Landscape of the Emergence of Life. Life 2017, 7, 27. [CrossRef]
- Hansma, H.G. Possible origin of life between mica sheets. Theor. Biol. 2010, 266, 175–188. [CrossRef]
- Cech, T.R. Self-splicing of group I introns. Rev. Biochem. 1990, 59, 543–568. [CrossRef]
- Ketting, R.F.; Fischer, S.E.J.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H.A. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in elegans. Genes Dev. 2001, 15, 2654–2659. [CrossRef]
- Altman, S. Structural biology. Nature 2000, 7, 827–828.
- Cech, T.R.; Zaug, A.J.; Grabowski, P.J. In Vitro splicing of the ribosomal RNA precursor of Tetrahymena: involvement of a guanosine nucleotides in the excision of the intervening sequence. Cell 1981, 27, 487–496. [CrossRef]
- Prosdocimia, F.; Jheeta, S.; Farias, S.T. Conceptual challenges for the emergence of the biological system: Cell theory and self-replication. Hypotheses 2018, 119, 79–83. [CrossRef]
- Hordijk, W.; Steel, M. Chasing the tail: The emergence of autocatalytic networks. Biosystems 2017, 152, 1–10. [CrossRef]
- Eigen, M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971, 58, 465–523. [CrossRef]
- Ameta, S.; Arsene, S.; Lehman, N.; Griffiths, A.D.; Nghe, P. Evolution using autocatalytic sets of RNA. Manuscript under preparation.
- Ameta, S.; Arsene, S.; Lehman, N.; Nghe, P.; Griffiths, A.D. (Conference Abstract) Autocatalytic sets of RNA replicators in origin of life. In Proceedings of the XVIIIth International Conference on the Origin of Life, San Diego, CA, USA, 16–21 July 2017; Volume 1967.
- Arsène, S.; Ameta, S.; Lehman, N.; Griffiths, A.D.; Nghe, P. Coupled catabolism and anabolism in autocatalytic RNA sets. Nucleic Acids Res. 2018, 46, 9660–9666. [CrossRef]
- Iqubal, M.A.; Sharma, R.; Jheeta, S. Thermal condensation of glycine and alanine on metal ferrite surface: primitive peptide bond formation scenario. Life 2017, 7, 15. [CrossRef]
- Sharma, R.; Iqubal, M.A.; Jheeta, S. Adsorption and Oxidation of Aromatic Amines on Metal(II) Hexacyanocobaltate(III) Complexes: Implication for Oligomerization of Exotic Aromatic Compounds. Inorganics 2017, 5, 18. [CrossRef]
- Kotakis, C. Non-coding RNAs’ partitioning in the evolution of photosynthetic organisms via energy transduction and redox signalling. RNA Biol. 2015, 12, 101–104. [CrossRef]
- Smith, D.R. RNA-Seq data: A goldmine for organelle research. Funct. Genomics 2013, 12, 454–456. [CrossRef]
- Kahana, A.; Lancet, D. Protobiotic Systems Chemistry Analyzed by Molecular Dynamics. Life 2019, 9, 38. [CrossRef]
- Lancet, D.; Zidovetzki, R.; Markovitch, O. Systems protobiology: origin of life in lipid catalytic networks. J. R. Soc. Interface 2018, 15. [CrossRef]
- Schopf, J.W.; Kitajima, K.; Spicuzza, M.J.; Kudryavtsev, A.B.; Valley, J.W. SIMS analyses of the oldest known assemblage of microfossils document their taxon-correlated carbon isotope compositions. Natl. Acad. Sci. USA 2018, 115, 53–58. [CrossRef]
- Jheeta, S. The Routes of Emergence of Life from LUCA during the RNA and Viral World: A Conspectus. Life 2015, 5, 1445–1453. [CrossRef]
- Koumandou, V.L.; Kossida, S. Evolution of the F0F1 ATP Synthase Complex in Light of the Patchy Distribution of Different Bioenergetic Pathways across Prokaryotes. PLOS Comput. Biol. 2014, 10, e1003821. [CrossRef]
- Agioutantis, P.; Koumandou, V.L. Bioenergetic diversity of the human gut microbiome. Meta Gene 2018, 16, 10–14. [CrossRef]
- Koumandou, V.L.; Kossida, S. Evolution of b-type cytochromes in prokaryotes. PeerJ PrePrints 2015. [CrossRef]
- Lang, A.S.; Westbye, A.B.; Beatty, J.T. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Rev. Virol. 2017, 4, 87–104. [CrossRef]
- Westbye, A.B.; Beatty, J.T.; Lang, A.S. Guaranteeing a captive audience: coordinated regulation of gene transfer agent (GTA) production and recipient capability by cellular regulators. Opin. Microbiol. 2017, 38, 122–129. [CrossRef]
- Ayre, D.C.C.; Elstner, M.; Smith, N.C.; Moores, E.S.; Hogan, A.M.; Christian, S.L. Dynamic regulation of CD24 expression and release of CD24-containing microvesicles in immature B cells in response to CD24 engagement. Immunology 2015, 146, 217–233. [CrossRef]
- Ayre, D.C.C.; Chute, I.C.; Joy, A.P.; Barnett, D.A.; Hogan, A.M.; Gruel, M.P.; Christian, S.L. CD24 induces changes to the surface receptors of B cell microvesicles with variable effects on their RNA and protein cargo. Rep. 2017, 7, 8642. [CrossRef]
- Sabi, R.; Tuller, T. Modelling and measuring intracellular competition for finite resources during gene expression. R. Soc. Interface 2019, 16. [CrossRef]
- Zur, H.; Tuller, T. Predictive biophysical modeling and understanding of the dynamics of mRNA translation and its evolution. Nucleic Acids Res. 2016, 44, 9031–9049. [CrossRef]
- Goz, E.; Zafrir, Z.; Tuller, T. Universal evolutionary selection for high dimensional silent patterns of information hidden in the redundancy of viral genetic code. Bioinformatics 2018, 34, 3241–3248. [CrossRef]
- Jheeta, S.; Ptasinska, S.; Sivaraman, B.; Mason, N.J. The irradiation of 1:1 mixture of ammonia:carbon dioxide ice at 30 K using 1 kev Electrons. Phys. Lett. 2012, 543, 208–212. [CrossRef]
- Jheeta, S.; Domaracka, A.; Ptasinska, S.; Mason, N.J. The irradiation of pure CH3OH and 1:1 mixture of NH3:CH3OH ices at 30 K using low energy electrons. Chem. Phys. Lett. 2013, 556, 359–364. [CrossRef]
- Jheeta, S. Final frontiers: The hunt for life elsewhere in the Universe. Space Sci. 2013, 348, 1–10. [CrossRef]
- Le Roy, L.; Briani, G.; Briois, C.; Cottin, H.; Fray, N.; Thirkell, L.; Poulet, G.; Hilchenbach, M. On the prospective detection of polyoxymethylene in comet 67P/Churyumov–Gerasimenko with the COSIMA instrument onboard Rosetta. Space Sci. 2012, 65, 83–92. [CrossRef]
- Kissel, J.; Altwegg, B.C.; Clark, L.; Colangeli, H.; Czempiel, S.; Eibl, J.; Engrand, C.; Fehringer, H.M.; Feuerbacher, B.; Fomenkova, M.; et al. COSIMA—High Resolution Time-of-Flight Secondary Ion Mass Spectrometer for the Analysis of Cometary Dust Particles onboard Rosetta. Space Sci. Rev. 2007, 128, 823–867. [CrossRef]
- Pleyer, H.L.; Strasdeit, H.; Fox, S. A Possible Prebiotic Ancestry of Porphyrin-Type Protein Cofactors. Life Evol. Biosph. 2018, 48, 347–371. [CrossRef]
- Fox, S.; Strasdeit, H. A Possible Prebiotic Origin on Volcanic Islands of Oligopyrrole-Type Photopigments and Electron Transfer Cofactors. Astrobiology 2013, 13, 578–595. [CrossRef]
- Fox, S.; Strasdeit, H. Inhabited or Uninhabited? Pitfalls in the Interpretation of Possible Chemical Signatures of Extraterrestrial Life. Microbiol. 2017, 8, 1622. [CrossRef]
- Schwieterman, E.W.; Kiang, N.Y.; Parenteau, M.N.; Harman, C.E.; DasSarma, S.; Fisher, T.M.; Arney, G.N.; Hilairy, E.; Hartnett, H.E.; Reinhard, C.T.; et al. Exoplanet Biosignatures: A Review of Remotely Detectable Signs of Life. Astrobiology 2018, 18. [CrossRef]
- Battistuzzi, F.U.; Feijao, A.; Hedges, S.B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 2004, 4, 44. [CrossRef]
- Georgiou, C.D. Functional properties of amino acid side chains as biomarkers of extraterrestrial life. Astrobiology 2018, 18, 1479–1496. [CrossRef]
- Georgiou, C.D.; Deamer, D.W. Lipids as universal biomarkers of extra-terrestrial life. Astrobiology 2014, 14, 541–549. [CrossRef]
- Ruiz-Mirazo, K.; Briones, C.; de la Escosura, A. Chemical roots of biological evolution: the origins of life as a process of development of autonomous functional systems. Open Biol. 2017, 7, 170050. [CrossRef]
- Saitta, M. From Computational Physics to the Origins of Life. In Life Sciences, Information Sciences; Thierry, G., Dominique, L., Marie-Christine, M., Jean-Charles, P., Eds.; Wiley: Hoboken, NJ, USA, 25 March 2018; Chapter [CrossRef]
- Uyama, T.; Hashimoto, J.; Kuzuhara, M.; Mayama, S.; Akiyama, E.; Currie, J.L.; Kudo, T.; Kusakabe, N.; Abe, L. The SEEDS High-Contrast Imaging Survey of Exoplanets around Young Stellar Objects. J. 2017, 153. [CrossRef]
- Uyama, T.; Tanigawa, T.; Hashimoto, J.; Tamura, M.; Aoyama, Y.; Brandt, T.D.; Ishizuka, M. Constraining Accretion Signatures of Exoplanets in the TW Hya Transitional Disk. J. 2017, 154, 90. [CrossRef]
- Uyama, T.; Hashimoto, J.; Muto, T.; Akiyama, E.; Dong, R.; de Leon, J.; Sakon, I.; Kudo, T.; Kusakabe, N.; Kuzuhara, M.; et al. Subaru/HiCIAO HKs Imaging of LKHa 330: Multi-band Detection of the Gap and Spiral-like Structures. Astron. J. 2018, 156, 63. [CrossRef]
- Wisłocka, A.M.; Kovacˇevic´, A.B.; Balbi, A. Comparative analysis of the influence of Sgr A* and nearby active galactic nuclei on the mass loss of known exoplanets. Astrophys. 2019, 624. [CrossRef]
- Luger, R.; Barnes, R.; Lopez, E.; Fortney, J.; Jackson, B.; Meadows, V. Habitable Evaporated Cores: Transforming Mini-Neptunes into Super-Earths in the Habitable Zones of M Dwarfs. Astrobiology 2015, 15, 57–88. [CrossRef]
- Valencia, D.; Ikoma, M.; Guillot, T.; Nettelmann, N. Composition and fate of short-period super-Earths: The case of CoRoTx7b. Astrophys. 2010, 516, A20. [CrossRef]
- Maccone, C. Energy of Extra-Terrestrial Civilizations according to Evo-SETI Theory. Acta Astronaut. 2018, 144, 202–213. [CrossRef]
- Maccone, C. Life Expectancy and Life Energy according to Evo-SETI Theory. J. Astrobiol. 2019, 189, 36–46. [CrossRef]
- Lazcano, A.; Miller, S.L. How long did it take for life to begin and evolve to cyanobacteria? Mol. Evol. 1994, 39, 546–554. [CrossRef]
- Forterre, P. Three RNA cells for ribosomal lineages and three DNAviruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. USA 2006, 10, 3669–3674. [CrossRef]


